Webinars du réseau "Phages.fr"
The Phages.fr network has launched in 2024 a webinars series – a rendez-vous to talk about these amazing viruses ! We propose a virtual stage (look at the replays here) to give the opportunity to the member of the network (priory given to Early Career Researchers) to unveil their groundbreaking results! This isn't just about presentations; it's about creating a 'friendly zone' where you can share and receive constructive feedback. These webinars provide the opportunity to exchange within the Phages.fr network, have better visibility of projects underway in the labs and allow participating members to identify potential collaborators and future partners they can contact if necessary. Mark your calendars for the last Wednesday of each month (may be flexible) at 1:30 p.m. Join us on Zoom for a 45-minutes session, with a 20-30 min talk followed by 15-20 minutes of discussion in English. Don't miss a beat!
Speaker application
Step into the limelight, share your discoveries and exchange ideas. Submit a short title/abstract of your talk. To do so, send an email to Anne Chevallereau, Yuvaraj Bhoobalan, Quentin Lamy-Besnier, Camille Kolenda, Emilie Cenraud
List of speakers in 2026
(see below for details) Watch in live the presentation / look at the replays here
-
January 28th: Nandita Sharma (Post-doc, Debarbieux lab, Institut Pasteur, Paris, France)
-
February 10th : Webinar France-Canada
-
March 25th: Youn le Cras (PhD student, Rocha lab, Institut Pasteur, Paris, France)
-
April 29th: Tristan Ferry (PU-PH, Hospices Civils de Lyon, UCBL, Lyon, France)
-
May 27th: Clarisse Plantady (PhD student, Chevallereau lab, MMSB, Lyon, France)
-
June 24th: Judith Sar (PhD student, LCB, Ansaldi lab, Marseille, France)
-
September 30th: Kevin Barthes (PhD student, Rousset lab, CIRI, Lyon, France)
-
October: Webinar France-Canada
-
November: Phages.fr annual conference in Toulouse
-
December 16th: tba
February 10th : Webinar France-Canada
Theme: Wine, cheese, and lactic acid bacteria: When Phages Show Up at the Table
view the presentations https://umontpellier-fr.zoom.us/j/2087814160?omn=98286518225
Program
-
Part 1 : Interactions between phages and bacteria in natural habitats subject to complex spatio-temporal stress patterns: the example of wine fermentation (presented by Claire Le Henaff-Le Marrec, Université de Bordeaux)
-
Part 2 : Guardians of yogurt and cheese: Meet the defense arsenal of Streptococcus thermophilus (presented by Sylvain Moineau, Université Laval, Québec)
-
Part 3 : Sweet receptors for phages in lactic acid bacteria (presented by Marie-Pierre Chapot-Chartier, INRAE, Jouy-en-Josas)
January 28th: Nandita Sharma
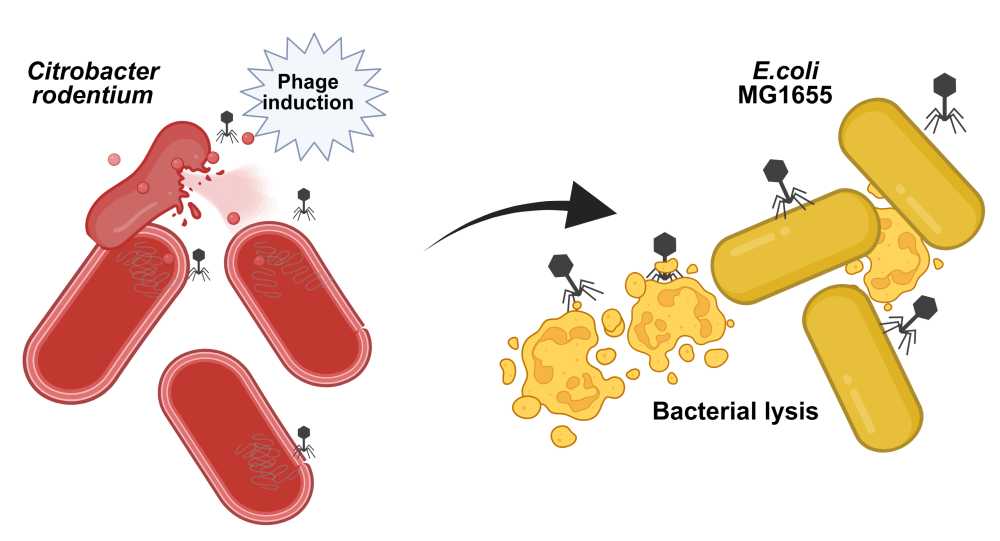
Understanding competition dynamics between Citrobacter rodentium and Escherichia coli in the gut microbiota
Nandita Sharma, post-doctorante à Bacteriophage, bacterium, host lab à Institut Pasteur, Paris, France.
Abstract
The gastrointestinal microbiota is a dynamic ecosystem in which bacteriophages play a role in killing bacteria from the “inside”, via prophage induction, and from the “outside”, by virulent bacteriophages. While prophage induction has been extensively studied in the laboratory through the lens of prophages encoding toxins, much less is known about the conditions and consequences of prophage induction in the intestinal microbiota. This study was initiated to investigate prophage induction during intestinal inflammation using gnotobiotic OMM12 mice infected by Citrobacter rodentium. Surprisingly, C. rodentium infection follows an original “peak and persistence” colonization pattern. Since the host immune response was insufficient for pathogen clearance, we introduced independently two murine commensal Escherichia coli strains (DN15 and Mt1B1) as well as the laboratory strain MG1655, all ahead of C. rodentium infection. Remarkably, DN15, but not Mt1B1 nor MG1655, substantially suppressed C. rodentium colonization. Comparative genomics and in vitro assays identified colicin Y, a plasmid-encoded antimicrobial peptide produced by DN15, as the key effector molecule responsible for inhibiting C. rodentium growth. Moreover, while DN15 and Mt1B1 loads remain constant, MG1655 loads decreased progressively, likely because of its susceptibility to two Citrobacter prophages (φNP and φSM). We leveraged this system by performing in vitro co-culture experiments with either wild-type or prophage-free mutants of C. rodentium and demonstrated that only φNP lysogenizes MG1655. Future plans include recording over time the in vivo induction of C. rodentium prophage and the detection of MG1655 lysogens. This study provides insights into the role of temperate phages as ecological modifiers of microbial communities in the gut.
List of speakers in 2025
(see below for details) Watch in live the presentation / look at the replays here
-
January 29th, 2025 - 1:30 p.m.: Pierre-Alexandre Pastouriaux (PhD student, CIRI, Lyon, France)
-
February 26th - 1:30 p.m.: Josie Elliott (post-doc, MMSB, Lyon, France)
-
March 26th - 1:30 p.m.: Julian Lopez (PhD student, MICALIS, Jouy-en-Josas, France)
-
April 30th - 1:30 p.m.: Eve Le Dauphin (PhD student, CEA-Leti Grenoble, France)
-
May 28th - 1:30 p.m.: Marianne Nicolas (post-doc, LCPME Nancy, France)
-
June 25th - 1:30 p.m.: Elsa Beurrier (PhD student, MIVEGEC, Montpellier, France)
-
September 23rd - 3:00 p.m.: webinars France/Canada (E. Rocha, I. Pasteur / F. Le Roux, U. Montreal / J. Nassimov, U. Waterloo)
-
October 29th - 1:30 p.m.: Benoît Lerouge (PhD student, INRAE, MICALIS, Jouy-en-Josas)
October 29th, 2025: Benoît Lerouge
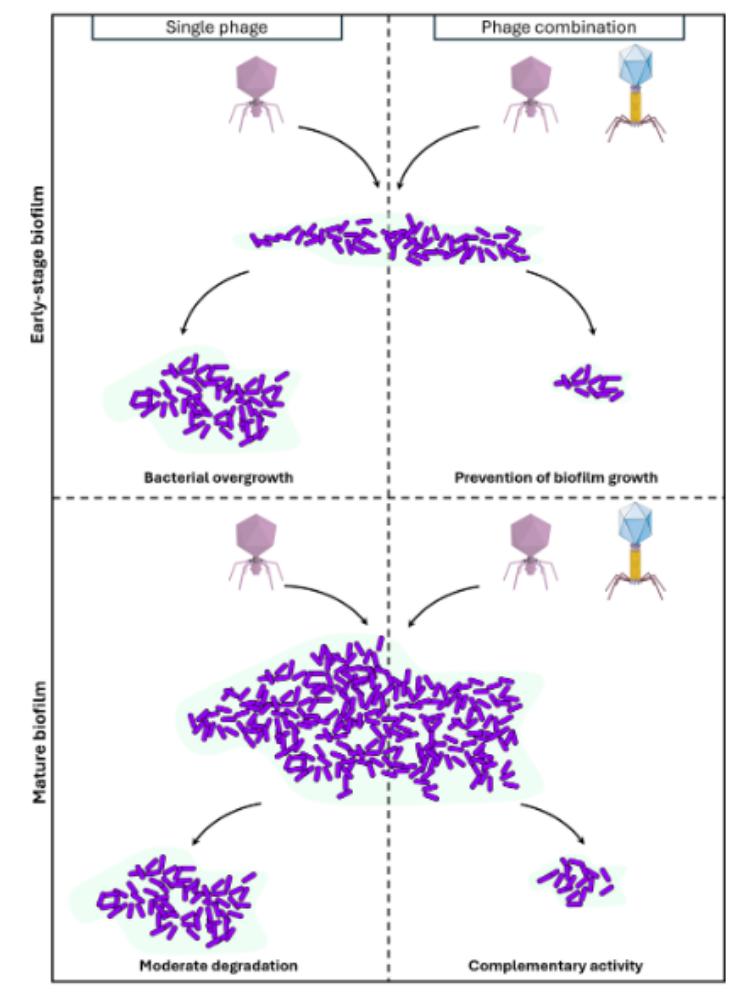
Activity of two Pseudomonas aeruginosa virulent phages on biofilms
Abstract
Pseudomonas aeruginosa is a ubiquitous Gram-negative bacterium responsible for various types of infections like bloodstream, urinary tract, eyes, wounds and chronic airways (especially in cystic fibrosis patients) infections. As a member of the ESKAPE group of pathogens, P. aeruginosa infections are difficult to treat due to antibiotic resistance and immune evasion mechanisms such as biofilm formation.
Biofilms are microbial communities encased in an extracellular matrix (ECM). In P. aeruginosa biofilms, ECM is mainly composed by three exopolysaccharides (EPS): alginate, Psl and Pel with different proportion depending on the strain, but also, by extracellular DNA and proteins. ECM forms a physical barrier which protects bacteria against the immune system and antimicrobial compounds like antibiotics.
Here, we explored the efficacity of two P. aeruginosa virulent phages on biofilms: a Pbunavirus LS1 which has O-antigen chain of the LPS as primary receptor, and a Bruynoghevirus LUZ24 whose receptor was unknown at the onset of this work. We firstly characterized the B. LUZ24 primary receptor as polysaccharide Psl, the main EPS in P. aeruginosa PAO1 biofilms. We next explored, by confocal laser microscopy, the ability of LUZ24 and LS1 phages to degrade PAO1 biofilms over a 48 hours period. Single phage application during the establishment phase of the biofilms (6 hours pre-culture) delayed biofilm growth by 18 hours, while the two phages combination prevented growth completely over 48 hours. In mature biofilms (16 hours pre-culture), at the end of the kinetics, single phage treatment led to a 2-fold reduction of viable bacteria, while the two phages combination led to a 4-fold reduction.
Overall, even if mature biofilms remain challenging to eradicate, these two phages present complementary killing activities, so that combining them is a promising strategy for phage therapy.
September 23rd, 2025 : Joint webinar between France and Canada
Ecology and molecular evolution of phage-bacteria relationships
-
3pm-3:30pm "The diversity of phage-bacteria interactions" by Eduardo Rocha (Pasteur Institute, Paris)
-
3:30pm-4pm "From isolation to validation: exploring phage–bacteria dynamics through spatiotemporal sampling and functional genetics" by Frédérique Le Roux (University of Montréal)
-
4pm-4 :30pm "Unravelling the “dark matter” of cyanophages: from protein structure to function" by Jozef Nassimov (University of Waterloo, Ontario)
-
4:30pm-5pm "Silent players in fermentation: Can phages affect cider quality?" by Marion Delmasso (University of Caen Normandie)
June 27th, 2025: Elsa Beurrier
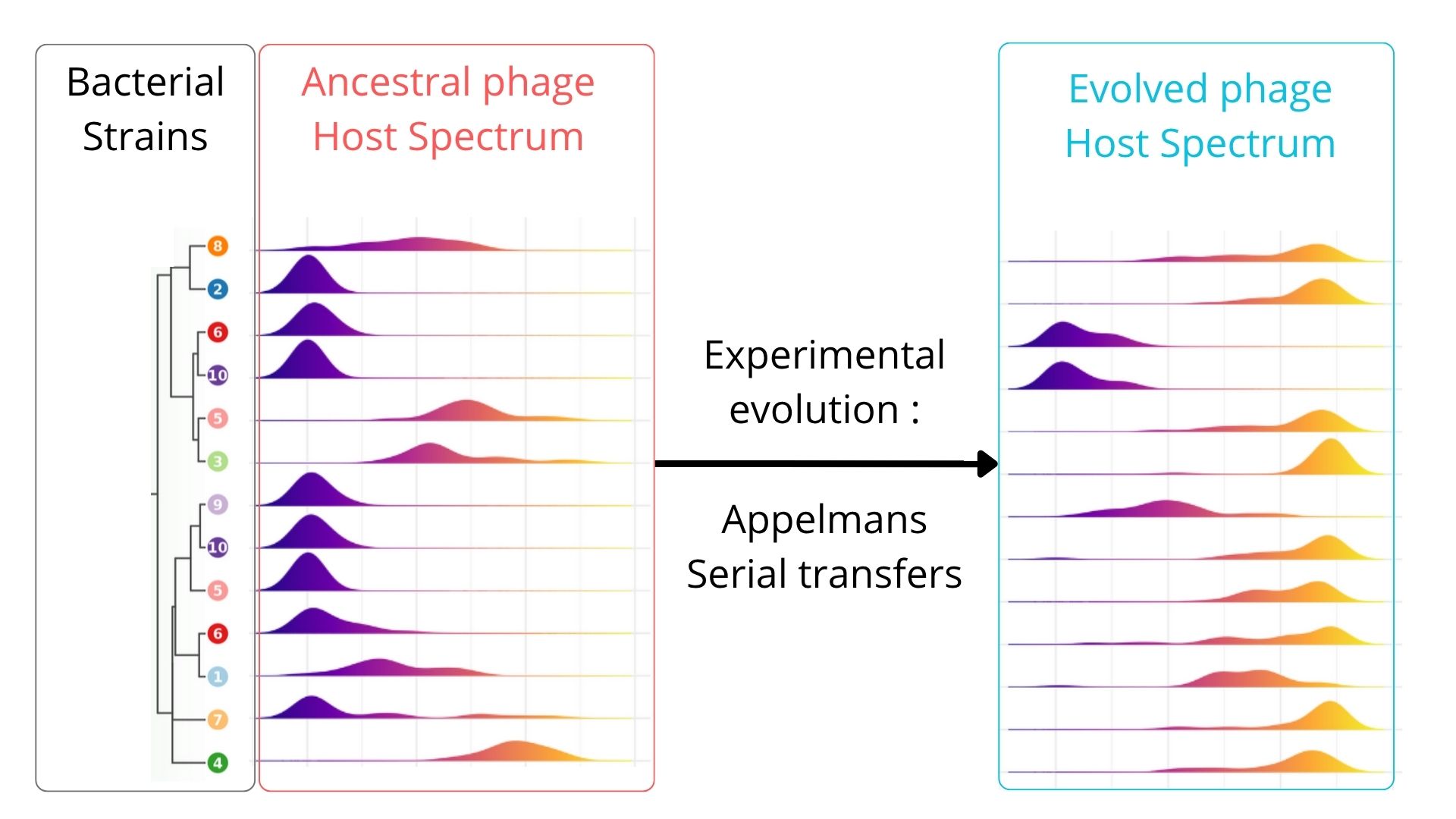
Experimental Evolution of a Tequatrovirus Bacteriophage Expands Host Range Against Carbapenem-Resistant Escherichia coli ST131
Elsa Beurrier, doctorante à MIVEGEC à Montpellier, France.
Abstract
The global spread of carbapenem-resistant Escherichia coli ST131 strains is a major public health issue, due to their increasing prevalence in community-acquired and nosocomial multidrug-resistant infections, and their high phenotypic and genotypic diversity. In this context, bacteriophage therapy might represent an interesting complementary strategy to control bacterial infections. On the one side, bacteriophages are specific and thus prevent the depletion of commensal bacteria. On the other side, such specificity impair the use of bacteriophages against multiple targeted pathogenic bacteria.
We have thus studied the evolutionary adaptation of a bacteriophage to an environment composed of multiple genetically diverse, non-coevolving clinical pathogenic isolates. The goal is to test the hypothesis: does evolving on a genetically diverse sub-collection enable infection of all isolates in the full collection?
We first isolated a virulent Tequatrovirus and tested its virulence over a collection of 134 bacterial isolates all attributed to 4 clades (A, B, C1 and C2) within the sequence type 131. Susceptible strains are scattered along the phylogenetic tree, but the C clade appears particularly resistant. The correlation between phylogenetic distance and phage susceptibility is currently being analyzed.
Based on Appelmans' evolution protocol (Burrowes 2019), we confronted the Tequatrovirus clonal population over 14 sequential passages to a bacterial sub-population composed of 46 bacterial strains of E. coli selected to be representative of the whole collection genomic diversity, based on the core-genome. This process was repeated over three independant phage populations.
Evolved phage populations show a significant broadening of the host spectrum by infecting 38/46 genotypes (82%) compared to 29/46 (63%) by the ancestral bacteriophage. 2/46 remained completely resistant (4.4%). We then assessed the activity of the evolved phage populations against the 89 additional E. coli ST131 genotypes not used during evolution. Interestingly, the evolved bacteriophages presented a significant larger host range to these genotypes by infecting 66/89 (i.e. 82%) while the ancestral population was infecting only 66/89 (i.e. 74%). 7/89 remained resistant to the evolved phage populations. No phylogeny signal could be found for the resistant isolated.
Our results suggest that using a phage isolated from the wild word lacks of efficacy against various compared to that same phage adapted to a subset of a bacterial genotypes. Directed evolution on a genetically varied bacterial population can significantly improve the efficacy and versatility of bacteriophages against multi-resistant strains of E. coli ST131.
May 28th, 2025: Marianne Nicolas
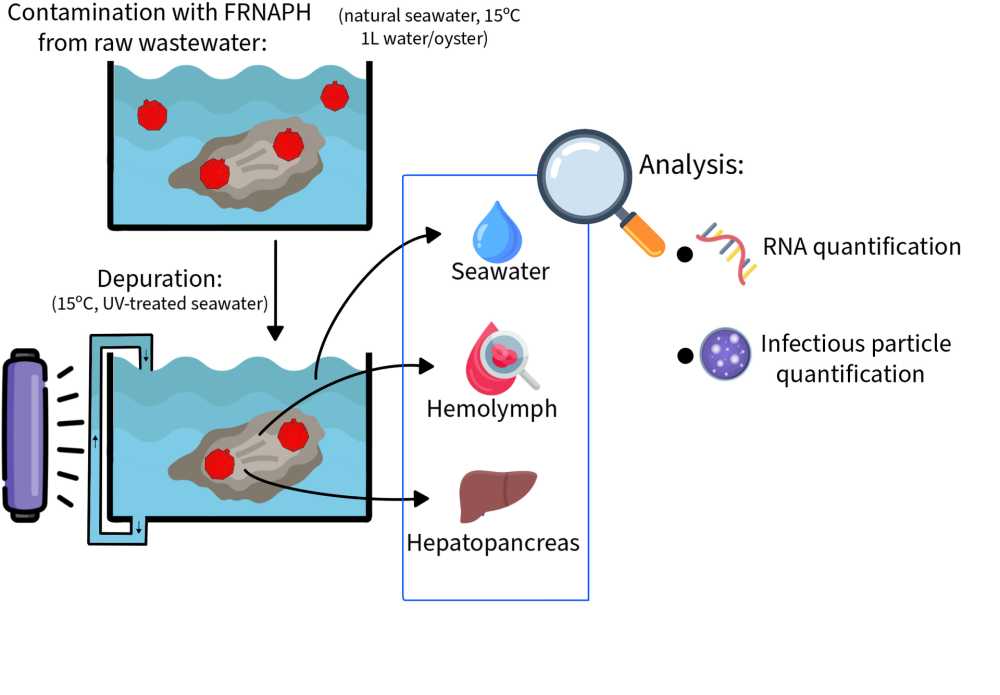
Study of viral depuration in oysters and characterization of F-specific RNA phages
Marianne Nicolas, post-doctorante au LCPME - UMR7564 CNRS à Nancy au laboratoire de "Microbiologie Environnementale" et à l'UMT Virocontrol chez ACTALIA à Saint Lô.
Abstract
Enteric viruses, including noroviruses, are a major cause of gastroenteritis associated with the consumption of contaminated shellfish. These viruses mainly originate from fecal pollution, often linked to wastewater discharges into the sea. Purification processes are commonly used in shellfish farming to reduce microbial contamination before commercialization. However, these methods are slow to eliminate viral genomes (1). In the absence of a culture method for noroviruses, only viral genome detection is currently possible. As no routine culture method is available for noroviruses, virological indicators have been proposed to assess the efficiency of oyster purification processes. In this context, the use of F-specific RNA bacteriophages (FRNAPH), which are naturally present in wastewater in high prevalence and concentration, structurally similar to enteric viruses, and easily cultivable in the laboratory, has been described (2,3).
The aim of the present study was to understand the mechanisms of viral particle inactivation and persistence in oysters during depuration. To this end, the presence of infectious and genomic FRNAPH was monitored in two oyster compartments (hepatopancreas and hemolymph) and in seawater.
Phage behavior in oysters was assessed in two depuration trials mimicking conditions of shellfish farms by using natural seawater continuously treated by UV light and maintained at about 15°C. FRNAPH used to contaminate oysters had been isolated from raw wastewater and replicated on E. coli to obtain sufficient concentrations for our experiments. The experimental contamination was performed by spiking the FRNAPH in the seawater and applying a contact time of 24h. FRNAPH were detected in the two compartments just after the accumulation phase. During the depuration, the FRNAPH were monitored in hepatopancreas and in hemolymph (over 14 days and 26 days in experiments 1 and 2, respectively). Infectious particles and genome of FRNAPH were predominantly detected in the hepatopancreas. A significant difference was observed between concentrations of infectious phages (-1.5 log10) and viral genomes (no change) in this organ (p < 0.01), highlighting that viral inactivation predominated on elimination and the central role of the hepatopancreas in viral retention. Although no release of phages from the hepatopancreas into the hemolymph was observed during depuration, this compartment be involved in viral inactivation. To explore this hypothesis, phage persistence in hemolymph was evaluated in vitro at 4°C. This experiment revealed prolonged stability of both infectious and genomic viral particles, suggesting that at this temperature, hemolymph does not contribute to phage inactivation.
These findings support the hypothesis that viral inactivation occurs within oysters, rather than elimination into the seawater during depuration. They also highlight the importance of simultaneously measuring infectious viruses and genomes to effectively assess purification processes. Further experiments are underway to reproduce phage persistence in hemolymph at a temperature similar to that used during depuration in shellfish farming.
1-Hartard C, Leclerc M, Rivet R, Maul A, Loutreul J, Banas S, Boudaud N, Gantzer C. F-specific RNA bacteriophages, especially members of subgroup II, should be reconsidered as good indicators of viral pollution of oysters. Dudley EG, editor. Appl Environ Microbiol. 2017 Dec;84(1):e01866-17.
2-Hodgson KR, Torok VA, Turnbull AR. Bacteriophages as enteric viral indicators in bivalve mollusc management. Food Microbiology. 2017 Aug;65:284–93.
3-Hartard C, Banas S, Rivet R, Boudaud N, Gantzer C. Rapid and sensitive method to assess human viral pollution in shellfish using infectious F-specific RNA bacteriophages: Application to marketed products. Food Microbiology. 2017 May;63:248–54.
April 30th, 2025: Eve Le Dauphin
Surface treatment to overcome phage adsorption during storage
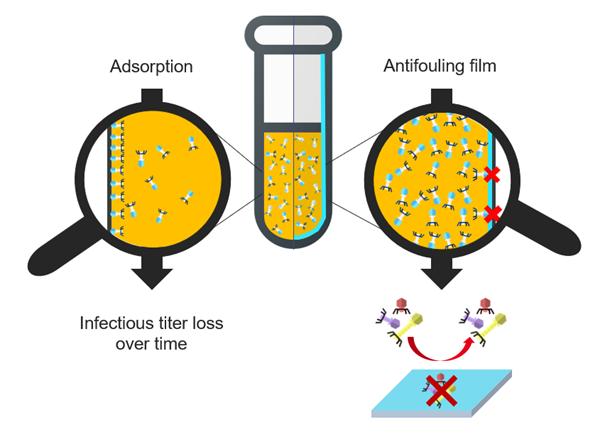
Courriel : Eve Le Dauphin, doctorante au CEA-Leti (Grenoble, France) sous la direction de Pierre MARCOUX, et Hippolyte DURAND.
Abstract
Bacterial resistance to antibiotics, also called antibioresistance, is increasing and “could cause 10 billion deaths each year by 2050” according to the World Health Organization [1]. Phagotherapy is a promising alternative to antibiotics, but as soon as bacteriophages are specific to bacterial hosts, a phage library at our disposal is needed to treat a maximum of bacterial infections. However, some preliminary studies [2,3] confirmed the essential role of the container material's surface properties which impacts phages concentration by decreasing by several decades the infectious titer over time.
The aim of the study is to develop vials with antibiofouling coating that prevents bacteriophages adsorption. We decided to focus on surface treatment by developing antibiofouling films on Glass and silicon substrates. PEG-like and zwitterion compounds are usually cites as having a well antifouling aptitude [4,5]. Once having measured the coating thickness by ellipsometry and evaluated its integrity during immersion, the film is tested as bacteriophages antifoulant. One method to conclude on the coating ability to be antibiofouling is the Quartz Cristal Microbalance. First, a buffer flow is used as reference, then phages with high concentration are introduced before washing with buffer again. Each coating is compared to a pristine substrate, so if the frequency signal comes back to reference frequency but the uncoated substrate does not, the coating is antifoulant. PEG-like and zwitterion films have been studied in that way and seems to be promising candidates to overcome bacteriophages adsorption.
Moreover, by doing QCM tests, we ask us the following question: do we adsorb only bacteriophages or are there other substances as Lipopolysaccharides ? During the presentation the contact between coatings and bacteriophages will be discussed using graphs obtained by QCM. Two methods used to respond to the LPS issue will be detailed by introducing bacteriophages purification and the measure of endotoxins level. Complementary analysis such as phages concentration in the post-QCM solution will support results.
References: [1] No Time to Wait: Securing the Future from Drug-Resistant Infections (29/04/20219). [2] Richter, Ł. et al. Adsorption of bacteriophages on polypropylene labware affects the reproducibility of phage research. Sci Rep 11, 7387 (2021). [3] O’Connell, L., Roupioz, Y. & Marcoux, P. R. Container material dictates stability of bacteriophage suspensions: Light scattering and infectivity measurements reveal mechanisms of infectious titre decay. Journal of Applied Microbiology (2022) doi:10.1111/jam.15581. [4] Hartig, T. et al. iCVD Polymer Thin Film Bio-Interface-Performance for Fibroblasts, Cancer-Cells, and Viruses Connected to Their Functional Groups and In Silico Studies. Advanced Materials Interfaces 11, 2300587 (2024). [5] Yang, R. & Gleason, K. K. Ultrathin Antifouling Coatings with Stable Surface Zwitterionic Functionality by Initiated Chemical Vapor Deposition (iCVD). Langmuir 28, 12266–12274 (2012).
March 26th, 2025: Julien Lopez
Bacteriophage lambda’s high mutation rate is not solely due to MMR inefficiency
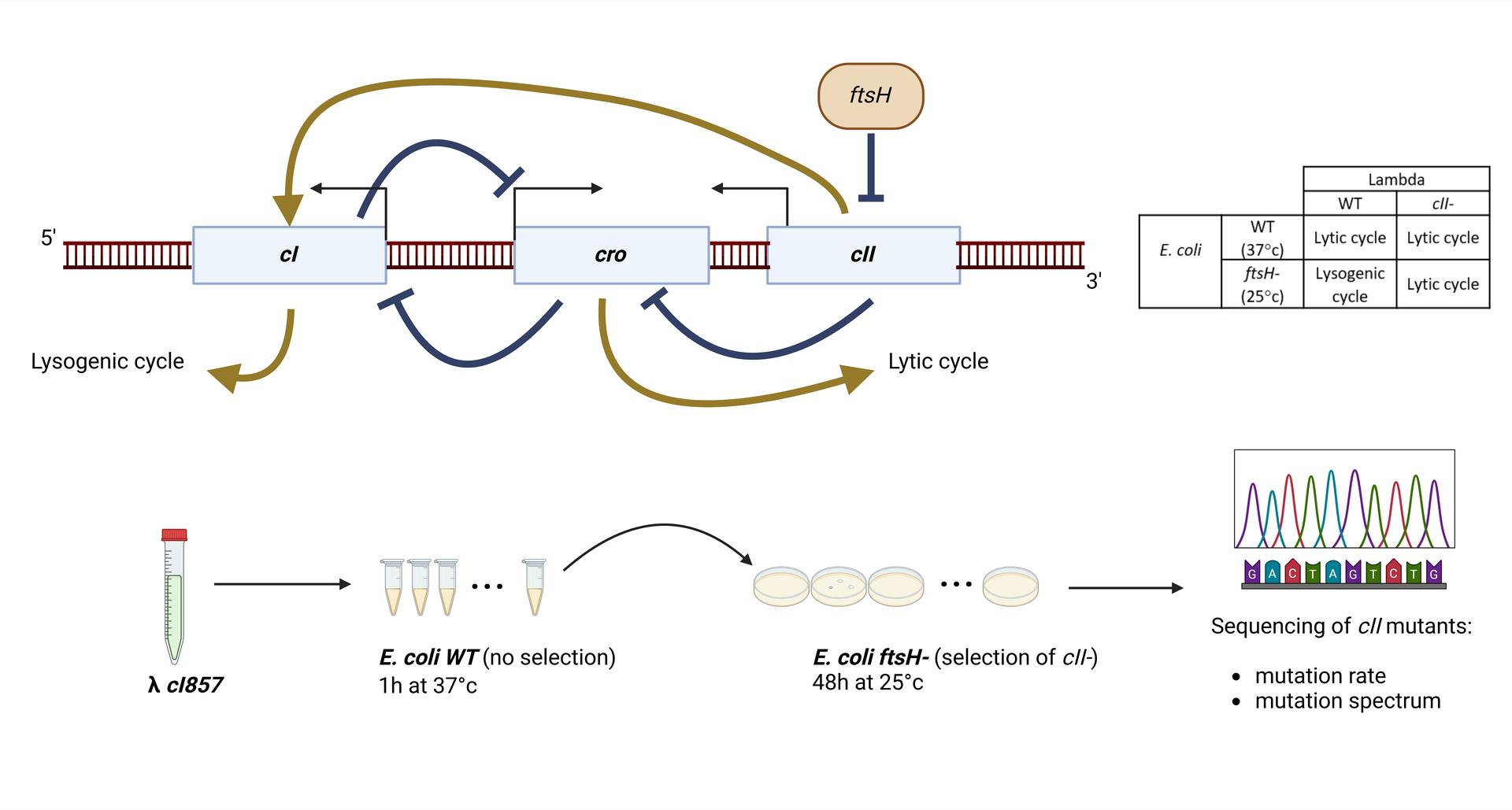
Courriel : Julien Lopez, doctorant dans le groupe "Mutagenesis in Single-Cells and Evolution (MuSE) au sein du laboratoire MICALIS Jouy-en-Josas, France.
Abstract
A single study¹ estimated that the mutation rate of lambda is 400-fold higher than that of its host, Escherichia coli (7.8x10-8 per base replicated for lambda vs. 2x10-10 for E. coli ²). Mutation rate is determined by replication fidelity and correction of errors (mainly by mismatch repair, MMR). Because E. coli DNA polymerase III is assumed to replicate both genomes³, the high mutation rate of lambda was supposed to be due to MMR inefficiency during infection⁴. This inefficiency was suspected to results from uncomplete adenine methylation of GATC sites in lambda’s DNA⁶ by Dam, as methylation of GATC is important for strand recognition and repair of mismatches by the MMR system.
In this work, we re-evaluated lambda mutation rate, and investigated the mechanisms of mutagenesis in lambda, notably the effect of Dam methylation. First, we estimated lambda mutation rate by a whole genome sequencing approach (Duplex Sequencing or DS⁽⁵⁾), that gave a slightly lower lambda mutation rate than previously estimated (1.5x10-8) and confirmed the poor efficiency of MMR on lambda DNA (2-fold increase for lambda vs. 150-fold increase for the host). In addition, we performed Fluctuation Assays⁷ (FA), that consist in determining the number of mutations in a target locus (here cII lambda gene) in a large number of parallel cultures, in different E. coli backgrounds. We found that the mutation rate of lambda with FA is close to that obtained with the DS experiment in the WT cells (1.8x10-8), but not in MutS E. coli cells (1.8x10-7). We also found that dam deletion increased lambda’s mutation rate to an intermediate level compared to the MutS deletion, as observed in E. coli. In addition, dam overexpression did not diminished lambda mutation rate, suggesting that a default of methylation does not explain lambda mutation rate. Finally, we determined the mutation spectrum of lambda by sequencing of a large number of cII mutants. This revealed that lambda mutation rate is different from that of either WT of MMR deficient E. coli ², and is not indicative of any known pathway of mutagenesis in E. coli.
Altogether these results indicate that lambda’s high mutation rate is not explained by MMR inactivity, as previously thought,but most likely by molecular mechanisms that modify the rate and the nature of Pol III replication errors on phage DNA. We are currently performing a Mutation Accumulation assay⁸, to confirm the mutation spectrum on the whole genome of lambda.
Bibliography
-
Dove, W. F. The genetics of the lambdoid phages. Annu. Rev. Genet 305–340 (1968).
-
Lee, H., Popodi, E., Tang, H. & Foster, P. L. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc. Natl. Acad. Sci. U. S. A. 109, (2012).
-
Zylicz, M., Ang, D., Liberek, K. & Georgopoulos, C. Initiation of λ DNA replication with purified host- and bacteriophage-encoded proteins: The role of the dnaK, dnaJ and grpE heat shock proteins. EMBO J. 8, 1601–1608 (1989).
-
Caillet-Fauquet, P., Maenhaut-Michel, G. & Radman, M. SOS mutator effect in E. coli mutants deficient in mismatch correction. EMBO J. 3, 707–712 (1984).
-
Kennedy, S. R. et al. Detecting ultralow-frequency mutations by Duplex Sequencing. Nat. Protoc. 9, 2586–2606 (2014).
-
Szyf, M., Avraham-Haetzni, K. & Reifman, A. DNA methylation pattern is determined by the intracellular level of the methylase. Proc. Natl. Acad. Sci. U. S. A. 81, 3278–3282 (1984).
-
Luria, S. E. & Delbrück, M. Mutations of Bacteria From Virus Sensitivity To Virus Resistance. Genetics 28, 491–511 (1943).
-
Halligan, D. L. & Keightley, P. D. Spontaneous mutation accumulation studies in evolutionary genetics. Annu. Rev. Ecol. Evol. Syst. 40, 151–172 (2009).
February 26th, 2025: Josie Elliott
Ecological Constraints Restrict the Benefits of Horizontally Acquired CRISPR-Cas Systems
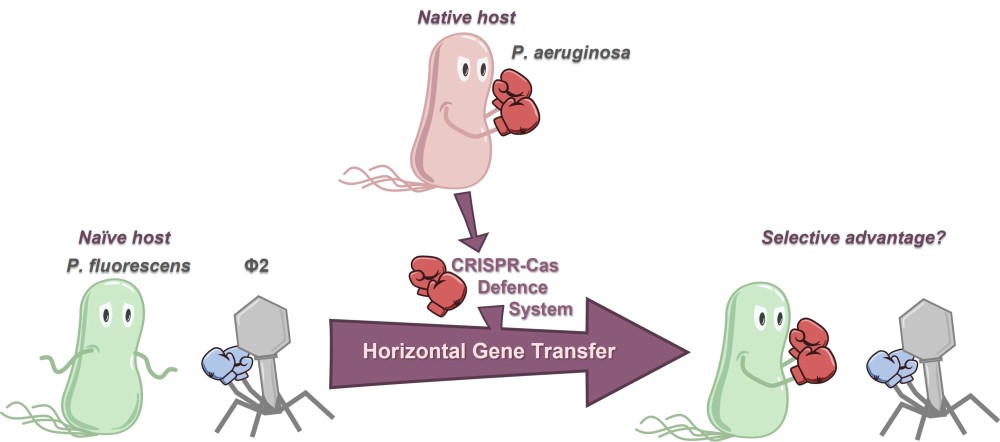
Courriel : Josie Elliott, post-doctorante dans le groupe d'Anne Chevallereau au laboratoire Molecular Microbiology and Structural Biochemistry, UMR5086 Lyon, France
Abstract
Acquiring a new defence system that can offer adaptive immunity against invading genetic elements (such as bacteriophage/phage) would be intuitively assumed to offer an evolutionary advantage to the host. Indeed, the ‘pan-immune model’ has proposed that possession of particular defence systems may be in a continual state of flux in microbial populations: allowing a reservoir of immune potential to be available without any single cell or strain having to carry them and bear the associated fitness costs. Furthermore, phage defence systems have been frequently associated with the hallmarks of horizontal gene transfer. Studies into adaptive-immunity-like CRISPR-Cas systems have supported the pan-immune model, with bioinformatic and experimental evidence for the frequent gain and loss of these systems. However, what is less well understood is the strength and potential conditional requirements to the evolutionary advantages offered to a host by a recently horizontally acquired CRISPR-Cas system. We experimentally modelled this ecological scenario by transferring the type I-F CRISPR-Cas system which is native to Pseudomonas aeruginosa to the naïve host Pseudomonas fluorescens. Although the CRISPR-Cas system is fully functional in the new host, protection against phage lysis is highly dependent on phage species and environment, and spacer acquisition against phage occurs at low frequency. This work shows that the ecological reality of successful selection after horizontal gene transfer events is nuanced. Therefore the limiting factor to successful sharing of defence systems in microbial populations might not only be rates of horizontal gene transfer, but also encountering ‘sweet spots’ of environmental conditions to produce robust selective benefits for hosts with the new defence system
January 29th, 2025: Pierre-Alexandre Pastouriaux
Impact of infection by virulent phages on the prophage induction in Staphylococcus aureus
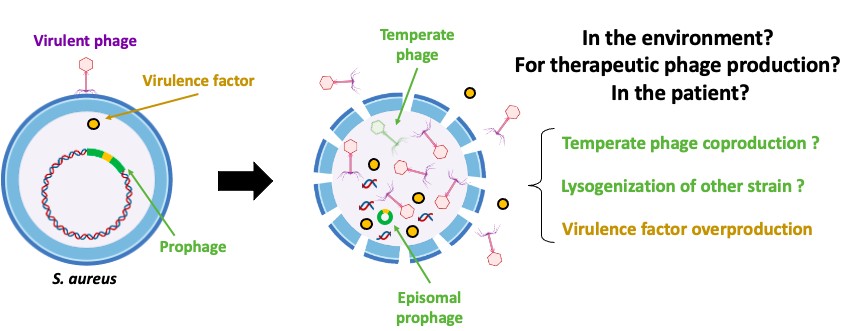
Courriel : Pierre-Alexandre Pastouriaux, Doctorant - CIRI website, Lyon, France
Abstract
Staphylococcus aureus is a major human pathogen, and the second leading cause of death associated with antibiotic resistance worldwide. Prophages play a key role in modulating the virulence and resistance of this bacterium, as well as in intra- and inter-species transfer of virulence/resistance genes. Understanding the phenomena of prophage induction is essential to better appreciate the environmental and epidemiological dynamics of S. aureus, especially in a context of renewed interest in phage therapy, which consists of using virulent phages to treat bacterial infections recalcitrant to conventional treatments. Indeed, the induction of prophages, either in the strain used to produce therapeutic phages or in the patient's strain during phage therapy, is a source of concerns for health authorities. However, few studies have focused on interactions between virulent phages, S. aureus and its prophages. In this context, the objective of this study is to evaluate the ability of virulent phages to induce prophages of S. aureus strains.
For this, the prophage content of S. aureus strains (n=172) was analyzed using PHASTER and BLAST. Then, 42 strains with various prophage contents (type Sa1: n=10; Sa2: n=16; Sa3: n=16) were infected at a MOI of 10, 10-3 or 10-5 with different virulent phages (n=4) belonging to the genera Silviavirus, Kayvirus or Rosenblumvirus. The induction of prophages leading to the production of temperate phages was then assessed by titration using the prophage-free S. aureus RN4220 strain which is susceptible to various phages, and by PCR and/or qPCR targeting virulence genes specific to prophage types Sa1int, Sa2int or Sa3int.
Overall, the results obtained show that, under these conditions, the presence of virulent phages at low MOI (10-3 and 10-5) does not appear to affect spontaneous prophage induction in S. aureus strains, nor to induce prophages that are not spontaneously induced. However, at MOI 10, they can trigger or amplify the spontaneous induction of specific prophages, including Eta/Sa1int and PVL/Sa2int prophages. These results highlight the ecological and clinical relevance of prophage dynamics in S. aureus. Further investigation will explore infections with multiple virulent phages and diverse environmental conditions. Additionally, lysogenization assays in the presence of virulent phages are planned to expand our understanding of these complex dynamics.
List of speakers in 2024
-
January 31st, 2024 : François Rousset (Post-doc Israel)
-
February 28th : Julian Bulssico (Post-doc LCB lab, CNRS Marseille)
-
March 27th: Florian Tesson (Doc. MDM lab, I. Pasteur Paris)
-
April 24th: Devon Conti (Doc. BBH lab, I. Pasteur Paris)
-
May 29th: Helena Shomar Monges (Post-doc. MDM lab, I. Pasteur Paris)
-
June 26th: Sol Vendrell-Fernández (Doc. GB lab, I. Pasteur Paris)
-
September 25th: Beatriz Beamud (Post-doc SB lab, I. Pasteur Paris)
-
October 30th: Benjamine Lapras (Doc. HCL, Lyon)
-
November 27th: Amandine Maurin (Doc at MIVEGEC lab, CNRS Montpellier)
-
December 18th : Morgane Illouz (post-doc at IRIM lab, CNRS Montpellier)
Dec 18th: Morgane Illouz
Role of trehalose polyphleates in the interactions between therapeutic phages and Mycobacterium abscessus
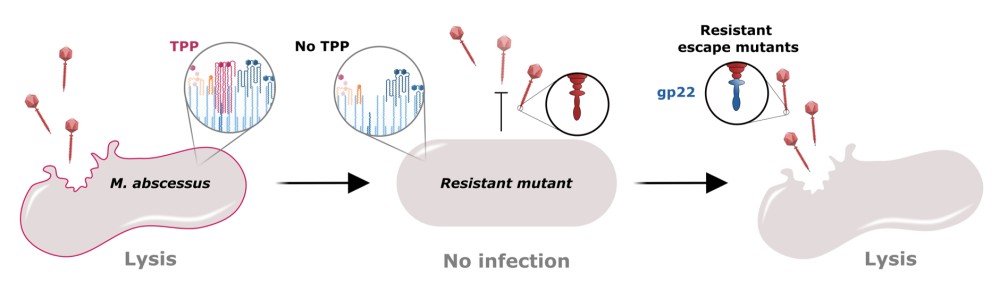
Morgane Illouz, Post-doc - UMR 9004 "Institut de Recherche en Infectiologie de Montpellier" IRIM website, Montpellier, France
Abstract
Cystic fibrosis patients are particularly susceptible to infections by the multidrug-resistant pathogen Mycobacterium abscessus (Mab), resulting in frequent therapeutic failures. Phage therapy, often in combination with antibiotics, has recently emerged as an alternative approach for the treatment of Mab pulmonary diseases. Despite its promise, the molecular mechanisms driving phage-host interactions and phage specificity in Mab remain poorly understood.
In this study, we investigated the role of mycobacterial surface components in the recognition and infection process of the therapeutic phage BPs targeting clinical Mab strains. Using a transposon mutant library, we identified phage-resistant Mab mutants, all carrying disruptions in genes involved in the biosynthesis of trehalose polyphleates (TPP), consisting of complex surface-associated lipids. In parallel, we also isolated spontaneous Mab mutants resistant to BPs with mutations in the TPP locus. Phage BPs failed to adsorb or infect TPP-deficient mutants, but these phenotypes were restored upon complementation of the disrupted genes.
To further elucidate this interaction, we employed fluorophages, enabling visualization and quantification of the infection process, to confirm the requirement of TPP in the adhesion process. Strikingly, we isolated BPs variants with mutations in the minor tail protein gp22, which conferred the ability to infect TPP-deficient Mab strains, illustrating a clear case of phage-bacteria co-evolution.
Our results uncover TPP as a novel receptor for mycobacteriophage adsorption and expand our understanding of early phage-host interaction mechanisms. Moreover, the identification of BPs variants with broader host range highlights their therapeutic potential against drug-resistant Mab, paving the way for the development of next-generation phage therapies against Mab diseases.
Nov 27th: Amandine Maurin
"This training is bound for glory": selection by experimental evolution of a phage with expanded host-range and increased virulence
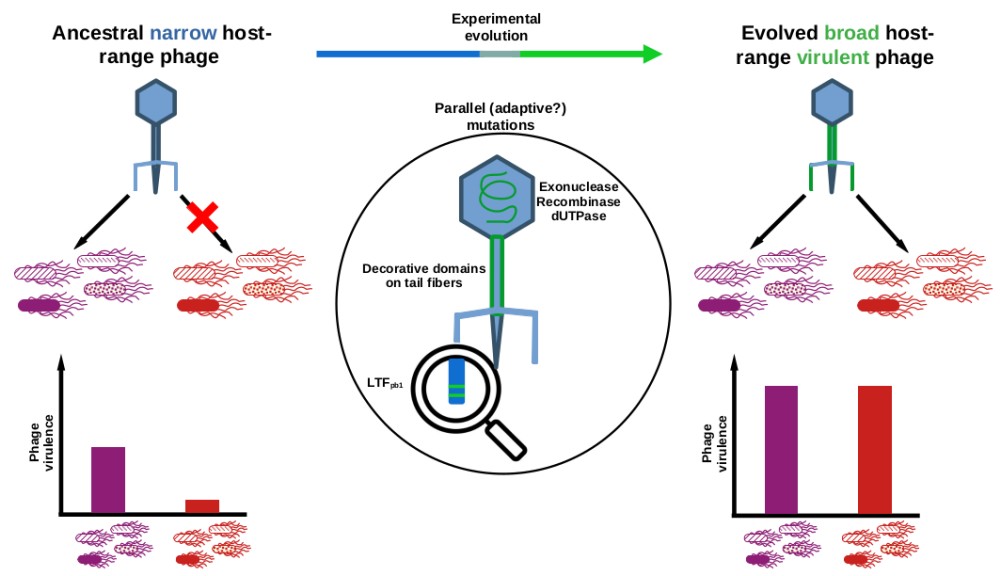
Amandine Maurin, PhD Student - UMR 5290 "Maladies infectieuses - Vecteurs, Ecologie, Génétique, Évolution et Contrôle" MIVEGEC website, Montpellier, France
Abstract
Parasites host-range expansion is predicted to evolve at the cost of lower mean fitness. We aimed at following the adaptive walks of a virulent phage (Tequintavirus) evolving in a spatially variable environment composed of four susceptible and four resistant isolates (Salmonella enterica serotype Tennessee, sequence types ST5018 and ST319 respectively). From a single ancestor, we evolved several independent populations through serial passages on the non-coevolving bacterial isolates following Appelmans' protocol. Evolved phage populations showed an expanded host-range and an increased virulence. Phage sequencing revealed multiple mutations appearing at the same loci (parallel mutations) in all populations, notably on exo- and endo-nuclease, dUTPase and caudal proteins. Interestingly, two parallel mutations present on the long tail fibre gene became fixed within the populations before other parallel mutations. Introduction of these two mutations by reverse-genetics into the ancestral genome expanded host-range but increased virulence only substantially, highlighting the effect of compensatory mutations.
October 30th: Benjamine Lapras
Rationalisation of the purification, formulation and packaging of anti-Staphylococcus aureus therapeutic phage suspensions for human use
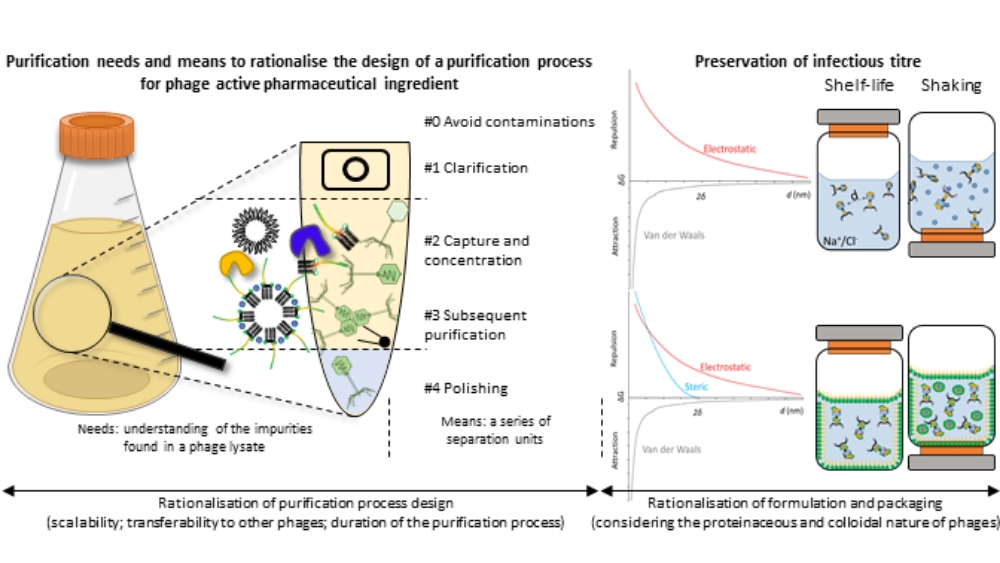
Benjamine Lapras, PhD Student - UMR 5305 Tissue Biology and Therapeutic Engineering Laboratory & FRIPHARM platform, Lyon Civil Hospices, Lyon
Abstract
The resurgence of phage therapy must overcome historical challenges intrinsic to phage suspensions, amongst which the long-term preservation of therapeutic concentrations of purified phage suspensions. To secure and accelerate the downstream processing (purification, formulation) of therapeutic phages, various properties of phages - and impurities - are rationalised to optimise the purification process, formulation and packaging, ensuring these methods can be applied to multiple phages and are compatible with human use. In this regard, the physicochemical properties of phages are considered to optimise the separation of the virions from bacterial impurities. The proteinaceous and colloidal (particles of 1 nm to 1 µm dispersed in a continuous medium) nature of phages is factored in to design and understand the thermodynamic interactions that lead to phage degradation mechanisms. When applied to anti-Staphylococcus aureus phage suspensions, this approach enabled the successful purification of one podovirus and three myoviruses. In parallel, a suspension medium was also developed and tested on one of the myovirus. It preserved the infectious titre of the phage under shelf-life conditions for a least a year (ongoing study) and for fourteen days under agitation. This work presents a theory-based methodology addressing pharmaco-technical challenges related to the purification and stabilisation of various phages to accelerate and secure phage development for therapeutic applications.
September 25th: Beatriz Beamud
Mapping factors of phage specificity: the case of restriction-modification systems
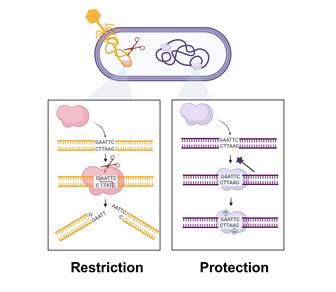
Beatriz Beamud, Post-doc in "Synthetic biology lab" (UMR6047, Intitut Pasteur, Paris)
Abstract
After successful recognition and adsorption to bacterial receptors, phages need to overcome a myriad of intracellular defenses. Among these, restriction-modification (RM) systems are ubiquitous in bacteria, leading to the degradation of foreign DNA that contains specific sequence motifs. In recent years, PacBio sequencing has enabled the identification of RM motifs by analyzing methylation patterns, revealing a great diversity of RM specificities. However, this technology has limitations, including ambiguous motif identification and high costs. To overcome this, we propose a method to detect restricted DNA sequences upon DNA transfer using random sequences (RandSeq). This approach has been validated with Escherichia coli strains with known restricted motifs and has revealed unknown specificities. Further, we compared restriction efficiency based on DNA delivery by conjugation or transduction, revealing RM systems to be more effective against phages. By identifying targeted DNA motifs, this method holds the potential to assist in the rational design of phages for therapeutic and biotechnological applications.
June 26th, 2024: Sol Vendrell-Fernández
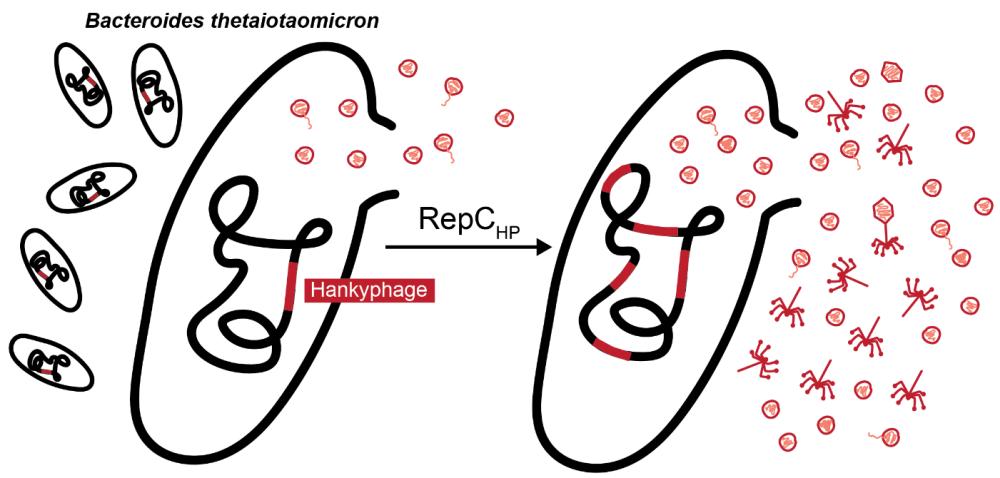
A CI-like repressor inhibits the Hankyphage lytic cycle in Bacteroides thetaiotaomicron
Sol Vendrell-Fernández, PhD student in "Genetics of Biofilms" (UMR6047, Intitut Pasteur, Paris)
Abstract
As one of the most prominent bacterial genera of the human gut, Bacteroides provide multiple nutritional benefits and contribute to the development of the host’s immune system. Similarly to other members the microbiota, Bacteroides engage in dynamic relationships with bacteriophages that impose selection pressures and ultimately contribute to community stability and adaptability. Temperate bacteriophages, especially, have been shown to contribute to bacterial fitness and survival through their versatile lifestyles, yet their lysogenic-lytic switch regulation remains scarcely studied to a molecular level in prominent non-model gut commensals. The Hankyphage is a broad-range temperate siphovirus that remarkably lysogenizes at least 28% species of the Bacteroides genus. In this study we investigated the Hankyphage lysogenic-lytic transition in the major gut symbiont B. thetaiotaomicron. We first observed that the Hankyphage and Bacteroides phylogenies were divergent, indicative of Hankyphage activity and horizontal transmission. Moreover, we showed that Hankyphage virions were spontaneously produced in absence of inducers, demonstrated that the phage replicates via replicative transposition and identified a repressor of phage transcription RepCHP capable of upregulating the lytic cycle. Additionally, we observed a generalized lack of plaque formation and secondary lysogenization that could be partly explained by intriguing phage structural defects. This study contributes to the understanding of the lysogenic-lytic switch of a prominent yet understudied bacteriophage of a relevant gut species.
May 29, 2024 : Helena Shomar
Genomics-driven discovery of a family of RiPPs that protect Actinobacteria from phage infection
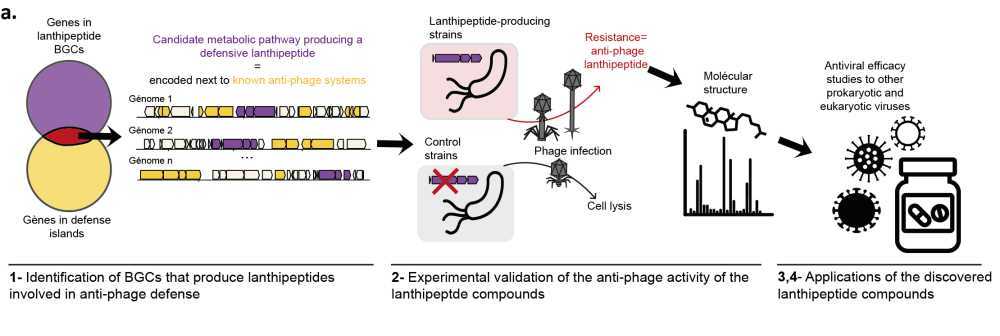
Helena Shomar, Post-doctoral researcher in "Molecular Diversity of Microbes" (MDM), Intitut Pasteur, Paris
Abstract
Bacteria produce a diverse array of natural products, to adapt to changing environments and stress. However, our understanding of the biological functions and ecological roles of the vast majority of these compounds remains limited. Genomic studies have unveiled the untapped metabolic potential of bacteria, with only 3% of natural products being characterized, and millions of molecules yet to be discovered. Recently, it emerged that a few known natural products allow bacteria to resist phage infection, but the prevalence of this defense strategy, called chemical defense, remains unclear. Here we use genomics and synthetic biology to uncover biosynthetic gene clusters that produce unknown natural products involved in anti-phage defense. We found that biosynthetic gene clusters that encode the production of a family of uncharacterized Ribosomally synthesized and post-translationally modified peptides (RiPPs) are often encoded near known anti-phage defense systems. Through heterologous expression in Streptomyces albus, we demonstrate experimentally the anti-phage activity of three representatives of this family of defensive RiPPs, present in hundreds of genomes of Actinobacteria. We further demonstrate the role of these defensive RiPPs in a native strain, allowing us to understand the regulation of their production. Finally, we delve into the anti-phage mechanism of action of these compounds. The discovery of defensive RiPPs paves the way for mining bacterial genomes for compounds involved in anti-phage defense, thus opening avenues for the development of new antiviral drugs derived from natural products.
April 24th, 2024: Devon Conti
The combined action of bacteriophages with a host-targeting molecule to clear the intestinal reservoir of multi-drug resistant Escherichia coli
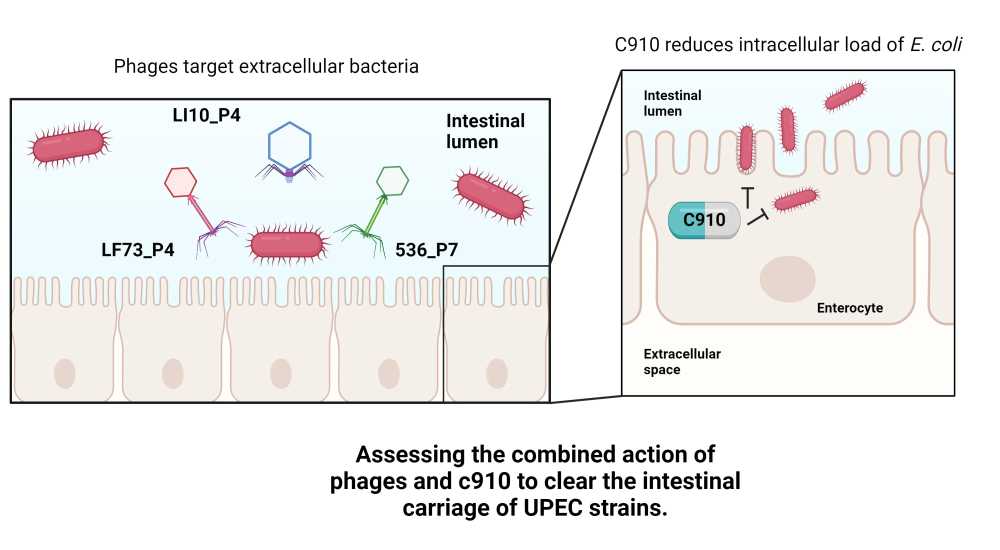
Devon Conti, PhD student in "Bacteriophage Bacterium Host", Pasteur Institute, Paris
Watch the replay here
Abstract
The multidrug resistant (MDR) Escherichia coli ST131 isolates are dominant extraintestinal pathogenic E. coli (ExPEC) with an increased capacity to colonize the gut, form intestinal reservoirs, and invade tissues. The CNF1 toxin expressed by these strains is a gut colonization factor involved in intestinal tissue invasion. Recently, the C910 compound was found to block CNF1-mediated host cell invasion(1).
The use of bacteriophages to reduce gut colonization of enteric pathogens has shown weak to moderate efficacy (2). We hypothesise that a double edge sword strategy based on the combination of bacteriophages with the C910 compound may enhance the clearance of enteric pathogens.
To setup the combined treatment, we first screened our phage collection and extensively assessed their activity in vitro to finally choose a cocktail of three genetically different phages. We found that the efficacy of this cocktail combined with the C910 molecule on epithelial cells infected with an ST131 isolate was synergistic, compared to individual treatments. Next, we used OMM12 mice, a gnotobiotic mouse model housing and transmitting a stable population of 12 bacteria and 11 induced prophages to evaluate the combined treatment in vivo 3. Following the characterization of the gut colonization profile of the ST131 isolate, including its capacity to form intracellular reservoir in the gut epithelium, we tested the combined treatment and observed an improved efficacy.
This work opens a new area of investigation to combine bacteriophages with molecules targeting host processes aiming at clearing intracellular reservoirs of bacterial pathogens.
-
Wu et al. iScience, 2022 (doi: 10.1016/j.isci.2022.104537)
-
Javaudin et al. Clin Microbiol Rev, 2021 (doi: 10.1128/CMR.00136-21)
-
Lamy-Besnier et al. Microbiome, 2023 (doi: 10.1186/s40168-023-01541-x)
March 27th, 2024 : Florian Tesson
The diversity of antiphage defense systems in prokaryotes

Florian Tesson, PhD student in "Molecular Diversity of Microbes" (MDM), Intitut Pasteur, Paris
Watch the replay here
Abstract
The evolutionary arms race between phages and bacteria led to the development of various defense mechanisms for bacteria and counter mechanisms for phages. Before 2015, only a few well-described defense systems were discovered. However, in recent years, the identification of "defense islands" and enhanced computational capacities, facilitated the discovery and validation of over a hundred novel antiphage defense systems.
While the number of different systems increased, the defense arsenal of a single bacterium was still unknown. To address this gap, we created DefenseFinder, a tool that systematically detects defense systems. Using this software, we analyzed more than 20,000 complete genomes to describe the different defense arsenals across the prokaryotic diversity. We investigate the diversity of systems present in each species and their relationship with mobile genetic elements.
Finally, while the discovery rate of defense systems increased, the discovery of anti-anti-phage defense encoded by phages and mobile genetic elements also increased. Thus, we developed a new tool, AntiDefenseFinder that systematically detects anti-defense systems a given genomes. We analyze AntiDefenseFinder results on more than 20,000 prokaryotic genomes and 10,000 phage genomes.
In summary, we offer an analysis of prokaryotic defense arsenal diversity and their link with mobile genetic elements alongside a new tool to detect viral counter-defense systems.
February 28th, 2024 : Julián Bulssico
Phage-Antibiotic synergy: single-phage/single-bacterium techniques to unveil the dynamics of phage propagation under antibiotic stress.
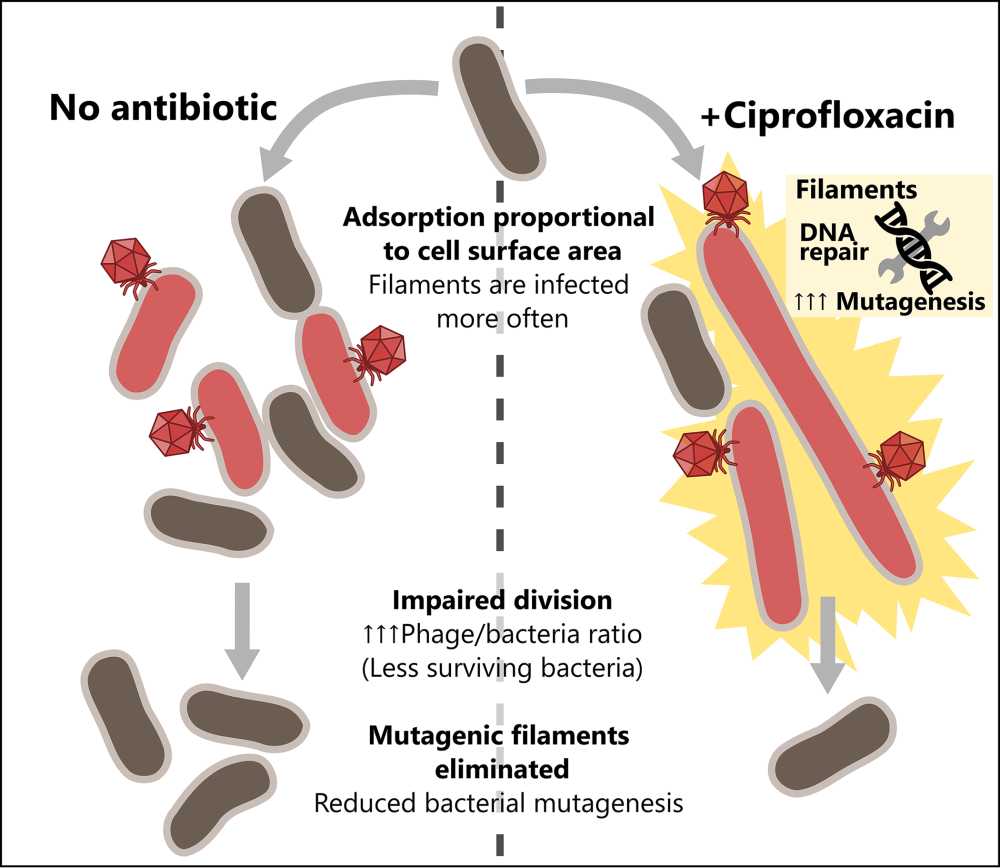
Julián Bulssico, Postdoctoral scientist - Laboratoire de Chimie Bactérienne, Marseille, France
Abstract
Antibiotic resistance is a major threat to public health, pushing the scientific community to develop alternative strategies. One of these is therapy combining bacteriophages and antibiotics, which is already administered in compassionate care. The combination of these two types of antibacterial, indeed, often displays a synergistic effect. This can be observed as enhanced phage propagation within a bacterial population in the presence of antibiotics. Despite the importance of such interactions, the molecular basis of synergy is far from being fully deciphered.
In our work, we characterised phage propagation in the presence of antibiotics. To achieve this, we developed powerful microscopy and image analysis techniques to visualize phage infection on E. coli with single-cell resolution. Our results suggest that synergy is remarkably conditioned by the structuration of the media in which phage and bacteria interact:
In well-mixed, liquid environments we assessed the impact of filamentation-inducing antibiotics on phage predation. We observed that impaired E. coli division enhances phage killing by phages HK620, T4, T5 and T7. Additionally, our novel tracking techniques allowed us to quantify the heterogeneity in phage infection. We observed that, due to their enlarged sizes, filaments are infected and lysed more often than regular-sized cells, which in turn limits SOS-mediated bacterial mutagenesis.
In semisolid media, we measured the impact of several synergistic antibiotics (ciprofloxacin, ceftazidime and mecillinam) on both phage T5 and T7, propagating in E. coli MG1655, expecting to find common effects that could explain enhanced phage propagation. We concluded that antibiotic-induced changes in bacterial morphology are crucial for the occurrence of synergy, and that two different altered shapes (spheroids and filaments) can lead to this effect.
Overall, we present new insights on the intricate interplay between phage, antibiotics, and bacteria, contributing to the characterization of epidemic propagation in bacterial populations suffering different types of stress, and highlighting the role of the spatial structuration of the system.
31 January 2024 : François Rousset
What bacterial defenses teach us about innate immunity?
François Rousset, Postdoctoral scientist - Weizmann Institute of Science
Watch the replay here
Abstract
Pathogens have fueled the diversification of intracellular defense strategies that collectively define cell-autonomous innate immunity. In bacteria, innate immunity is manifested by a broad arsenal of defense systems that provide protection against phages. The complexity of the bacterial immune repertoire has only been realized recently and is now suggesting that innate immunity has commonalities across the tree of life: many components of eukaryotic innate immunity are found in bacteria where they protect against phages. Here, I summarize recent findings on the conservation of innate immune pathways between prokaryotes and eukaryotes. I show that bacterial defense mechanisms can in turn catalyze the discovery of novel molecular players of eukaryotic innate immunity. We recently described ATP nucleosidases, immune effectors which cleave ATP molecules into adenine and ribose-5'-triphosphate during phage infection, thereby depriving phages of energy. Using phylogenetic analyses, we found that the immune ATP nucleosidase domain is found in a variety of eukaryotic organisms ranging from fungi to corals and insects, where it is embedded in a diverse set of proteins with a typical immune architecture. Taken together, our findings suggest that ATP degradation represents a novel mechanism of innate immunity that is conserved across the tree of life, highlighting the potential of bacterial defenses to expand our knowledge of eukaryotic immunity.