Webinars du réseau "Phages.fr"
The Phages.fr network has launched in 2024 a webinars series – a rendez-vous to talk about these amazing viruses ! We propose a virtual stage (look at the replays here) to give the opportunity to the member of the network (priory given to Early Career Researchers) to unveil their groundbreaking results! This isn't just about presentations; it's about creating a 'friendly zone' where you can share and receive constructive feedback. These webinars provide the opportunity to exchange within the Phages.fr network, have better visibility of projects underway in the labs and allow participating members to identify potential collaborators and future partners they can contact if necessary.
Mark your calendars for the last Wednesday of each month (may be flexible) at 1:30 p.m. Join us on Zoom for a 45-minutes session, with a 20-30 min talk followed by 15-20 minutes of discussion in English. Don't miss a beat! Subscribe now to stay informed about upcoming bacteriophage webinars. To do so, send an email to sympa@services.cnrs.fr with "SUBSCRIBE webinars-phages-fr Prénom Nom " in the subject line. Replace ‘Prénom Nom’ by your first name and surname. The body of the email remains blank. To subscribe, click to the following link: Phages.fr_Webinars
Speaker application
Step into the limelight, share your discoveries and exchange ideas. Submit a short title/abstract of your talk. To do so, send an email to Anne Chevallereau, Yuvaraj Bhoobalan, Quentin Lamy-Besnier, Camille Kolenda, Emilie Cenraud
List of speakers in 2024
(see below for details) Watch in live the presentation here look at the replays here
-
January 31st, 2024 : François Rousset (Post-doc Israel)
-
February 28th : Julian Bulssico (Post-doc LCB lab, CNRS Marseille)
-
March 27th: Florian Tesson (Doc. MDM lab, I. Pasteur Paris)
-
April 24th: Devon Conti (Doc. BBH lab, I. Pasteur Paris)
-
May 29th: Helena Shomar Monges (Post-doc. MDM lab, I. Pasteur Paris)
-
June 26th: Sol Vendrell-Fernández (Doc. GB lab, I. Pasteur Paris)
-
September 25th: Beatriz Beamud (Post-doc SB lab, I. Pasteur Paris)
-
October 30th: Benjamine Lapras (Doc. HCL, Lyon)
-
November 27th: Amandine Maurin (Doc MIVEGEC lab, CNRS Montpellier)
-
December 18th : Morgane Illouz (IRIM, CNRS Montpellier)
September 25th: Beatriz Beamud
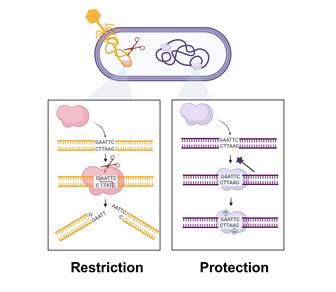
Mapping factors of phage specificity: the case of restriction-modification systems
Beatriz Beamud, Post-doc in "Synthetic biology lab" (UMR6047, Intitut Pasteur, Paris)
Abstract
After successful recognition and adsorption to bacterial receptors, phages need to overcome a myriad of intracellular defenses. Among these, restriction-modification (RM) systems are ubiquitous in bacteria, leading to the degradation of foreign DNA that contains specific sequence motifs. In recent years, PacBio sequencing has enabled the identification of RM motifs by analyzing methylation patterns, revealing a great diversity of RM specificities. However, this technology has limitations, including ambiguous motif identification and high costs. To overcome this, we propose a method to detect restricted DNA sequences upon DNA transfer using random sequences (RandSeq). This approach has been validated with Escherichia coli strains with known restricted motifs and has revealed unknown specificities. Further, we compared restriction efficiency based on DNA delivery by conjugation or transduction, revealing RM systems to be more effective against phages. By identifying targeted DNA motifs, this method holds the potential to assist in the rational design of phages for therapeutic and biotechnological applications.
June 26th, 2024: Sol Vendrell-Fernández
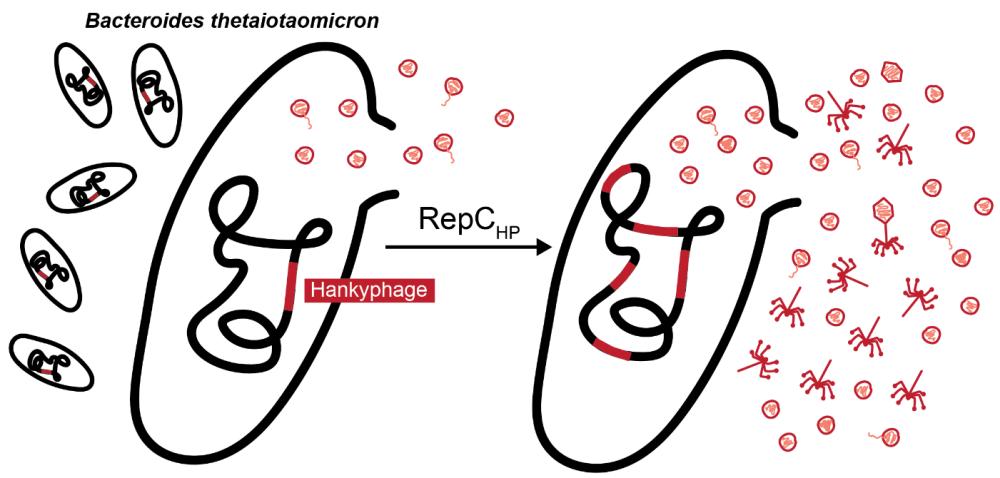
A CI-like repressor inhibits the Hankyphage lytic cycle in Bacteroides thetaiotaomicron
Sol Vendrell-Fernández, PhD student in "Genetics of Biofilms" (UMR6047, Intitut Pasteur, Paris)
Abstract
As one of the most prominent bacterial genera of the human gut, Bacteroides provide multiple nutritional benefits and contribute to the development of the host’s immune system. Similarly to other members the microbiota, Bacteroides engage in dynamic relationships with bacteriophages that impose selection pressures and ultimately contribute to community stability and adaptability. Temperate bacteriophages, especially, have been shown to contribute to bacterial fitness and survival through their versatile lifestyles, yet their lysogenic-lytic switch regulation remains scarcely studied to a molecular level in prominent non-model gut commensals. The Hankyphage is a broad-range temperate siphovirus that remarkably lysogenizes at least 28% species of the Bacteroides genus. In this study we investigated the Hankyphage lysogenic-lytic transition in the major gut symbiont B. thetaiotaomicron. We first observed that the Hankyphage and Bacteroides phylogenies were divergent, indicative of Hankyphage activity and horizontal transmission. Moreover, we showed that Hankyphage virions were spontaneously produced in absence of inducers, demonstrated that the phage replicates via replicative transposition and identified a repressor of phage transcription RepCHP capable of upregulating the lytic cycle. Additionally, we observed a generalized lack of plaque formation and secondary lysogenization that could be partly explained by intriguing phage structural defects. This study contributes to the understanding of the lysogenic-lytic switch of a prominent yet understudied bacteriophage of a relevant gut species.
May 29, 2024 : Helena Shomar
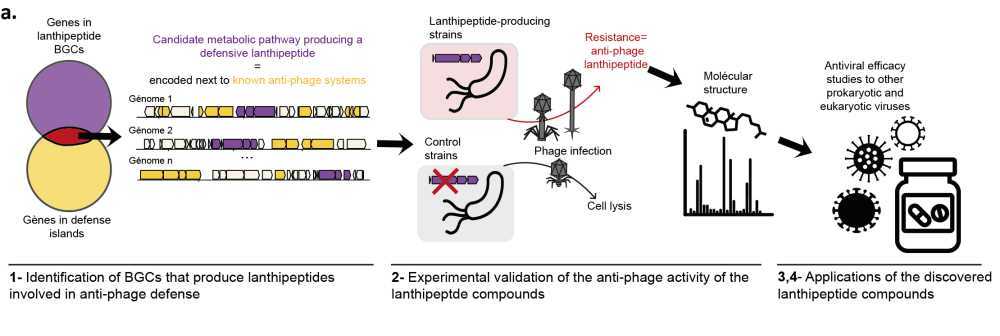
Genomics-driven discovery of a family of RiPPs that protect Actinobacteria from phage infection
Helena Shomar, Post-doctoral researcher in "Molecular Diversity of Microbes" (MDM), Intitut Pasteur, Paris
Abstract
Bacteria produce a diverse array of natural products, to adapt to changing environments and stress. However, our understanding of the biological functions and ecological roles of the vast majority of these compounds remains limited. Genomic studies have unveiled the untapped metabolic potential of bacteria, with only 3% of natural products being characterized, and millions of molecules yet to be discovered. Recently, it emerged that a few known natural products allow bacteria to resist phage infection, but the prevalence of this defense strategy, called chemical defense, remains unclear. Here we use genomics and synthetic biology to uncover biosynthetic gene clusters that produce unknown natural products involved in anti-phage defense. We found that biosynthetic gene clusters that encode the production of a family of uncharacterized Ribosomally synthesized and post-translationally modified peptides (RiPPs) are often encoded near known anti-phage defense systems. Through heterologous expression in Streptomyces albus, we demonstrate experimentally the anti-phage activity of three representatives of this family of defensive RiPPs, present in hundreds of genomes of Actinobacteria. We further demonstrate the role of these defensive RiPPs in a native strain, allowing us to understand the regulation of their production. Finally, we delve into the anti-phage mechanism of action of these compounds. The discovery of defensive RiPPs paves the way for mining bacterial genomes for compounds involved in anti-phage defense, thus opening avenues for the development of new antiviral drugs derived from natural products.
April 24th, 2024: Devon Conti
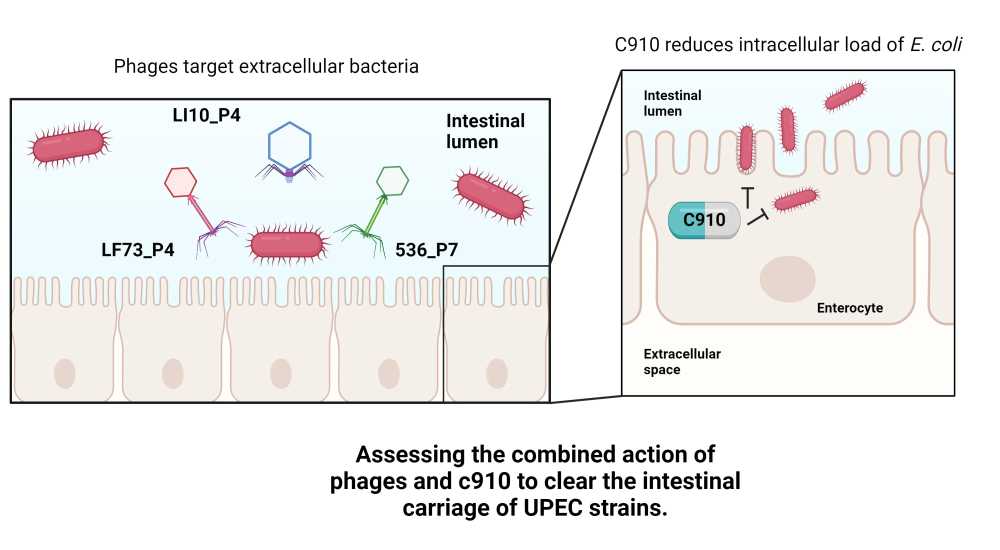
The combined action of bacteriophages with a host-targeting molecule to clear the intestinal reservoir of multi-drug resistant Escherichia coli
Devon Conti, PhD student in "Bacteriophage Bacterium Host", Pasteur Institute, Paris
Watch the replay here
Abstract
The multidrug resistant (MDR) Escherichia coli ST131 isolates are dominant extraintestinal pathogenic E. coli (ExPEC) with an increased capacity to colonize the gut, form intestinal reservoirs, and invade tissues. The CNF1 toxin expressed by these strains is a gut colonization factor involved in intestinal tissue invasion. Recently, the C910 compound was found to block CNF1-mediated host cell invasion(1).
The use of bacteriophages to reduce gut colonization of enteric pathogens has shown weak to moderate efficacy (2). We hypothesise that a double edge sword strategy based on the combination of bacteriophages with the C910 compound may enhance the clearance of enteric pathogens.
To setup the combined treatment, we first screened our phage collection and extensively assessed their activity in vitro to finally choose a cocktail of three genetically different phages. We found that the efficacy of this cocktail combined with the C910 molecule on epithelial cells infected with an ST131 isolate was synergistic, compared to individual treatments. Next, we used OMM12 mice, a gnotobiotic mouse model housing and transmitting a stable population of 12 bacteria and 11 induced prophages to evaluate the combined treatment in vivo 3. Following the characterization of the gut colonization profile of the ST131 isolate, including its capacity to form intracellular reservoir in the gut epithelium, we tested the combined treatment and observed an improved efficacy.
This work opens a new area of investigation to combine bacteriophages with molecules targeting host processes aiming at clearing intracellular reservoirs of bacterial pathogens.
-
Wu et al. iScience, 2022 (doi: 10.1016/j.isci.2022.104537)
-
Javaudin et al. Clin Microbiol Rev, 2021 (doi: 10.1128/CMR.00136-21)
-
Lamy-Besnier et al. Microbiome, 2023 (doi: 10.1186/s40168-023-01541-x)
March 27th, 2024 : Florian Tesson
The diversity of antiphage defense systems in prokaryotes

Florian Tesson, PhD student in "Molecular Diversity of Microbes" (MDM), Intitut Pasteur, Paris
Watch the replay here
Abstract
The evolutionary arms race between phages and bacteria led to the development of various defense mechanisms for bacteria and counter mechanisms for phages. Before 2015, only a few well-described defense systems were discovered. However, in recent years, the identification of "defense islands" and enhanced computational capacities, facilitated the discovery and validation of over a hundred novel antiphage defense systems.
While the number of different systems increased, the defense arsenal of a single bacterium was still unknown. To address this gap, we created DefenseFinder, a tool that systematically detects defense systems. Using this software, we analyzed more than 20,000 complete genomes to describe the different defense arsenals across the prokaryotic diversity. We investigate the diversity of systems present in each species and their relationship with mobile genetic elements.
Finally, while the discovery rate of defense systems increased, the discovery of anti-anti-phage defense encoded by phages and mobile genetic elements also increased. Thus, we developed a new tool, AntiDefenseFinder that systematically detects anti-defense systems a given genomes. We analyze AntiDefenseFinder results on more than 20,000 prokaryotic genomes and 10,000 phage genomes.
In summary, we offer an analysis of prokaryotic defense arsenal diversity and their link with mobile genetic elements alongside a new tool to detect viral counter-defense systems.
February 28th, 2024 : Julián Bulssico
Phage-Antibiotic synergy: single-phage/single-bacterium techniques to unveil the dynamics of phage propagation under antibiotic stress.
Julián Bulssico, Postdoctoral scientist - Laboratoire de Chimie Bactérienne, Marseille, France
Abstract
Antibiotic resistance is a major threat to public health, pushing the scientific community to develop alternative strategies. One of these is therapy combining bacteriophages and antibiotics, which is already administered in compassionate care. The combination of these two types of antibacterial, indeed, often displays a synergistic effect. This can be observed as enhanced phage propagation within a bacterial population in the presence of antibiotics. Despite the importance of such interactions, the molecular basis of synergy is far from being fully deciphered.
In our work, we characterised phage propagation in the presence of antibiotics. To achieve this, we developed powerful microscopy and image analysis techniques to visualize phage infection on E. coli with single-cell resolution. Our results suggest that synergy is remarkably conditioned by the structuration of the media in which phage and bacteria interact:
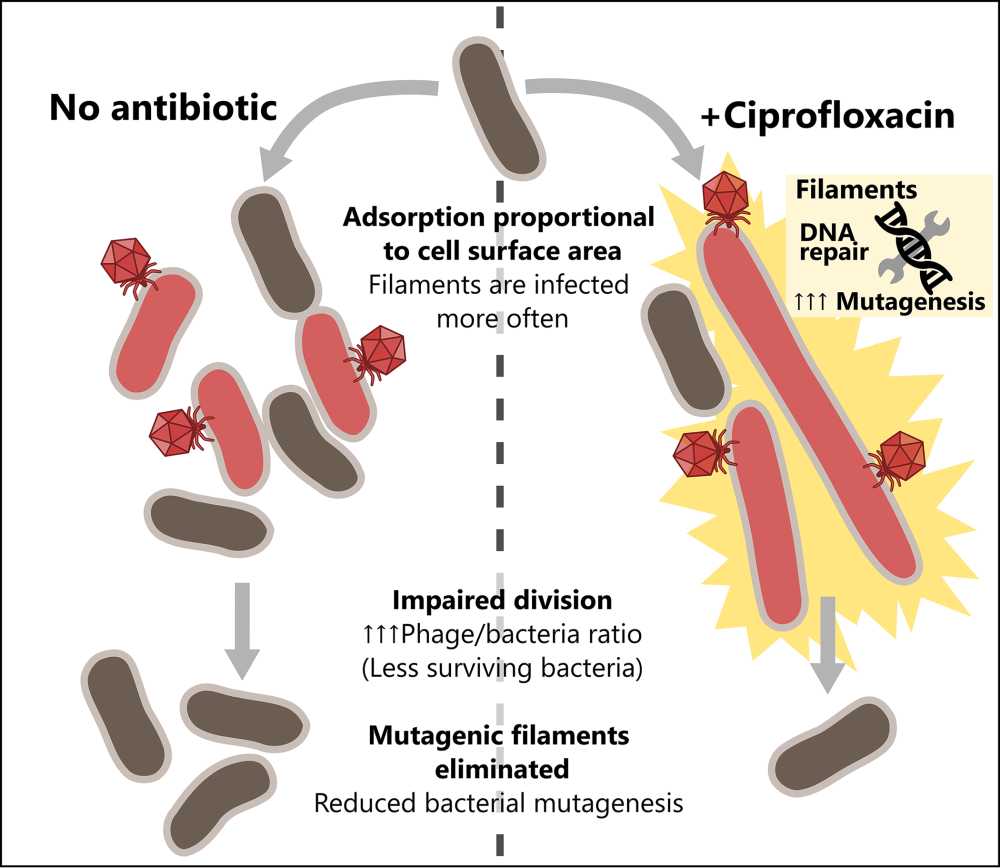
In well-mixed, liquid environments we assessed the impact of filamentation-inducing antibiotics on phage predation. We observed that impaired E. coli division enhances phage killing by phages HK620, T4, T5 and T7. Additionally, our novel tracking techniques allowed us to quantify the heterogeneity in phage infection. We observed that, due to their enlarged sizes, filaments are infected and lysed more often than regular-sized cells, which in turn limits SOS-mediated bacterial mutagenesis.
In semisolid media, we measured the impact of several synergistic antibiotics (ciprofloxacin, ceftazidime and mecillinam) on both phage T5 and T7, propagating in E. coli MG1655, expecting to find common effects that could explain enhanced phage propagation. We concluded that antibiotic-induced changes in bacterial morphology are crucial for the occurrence of synergy, and that two different altered shapes (spheroids and filaments) can lead to this effect.
Overall, we present new insights on the intricate interplay between phage, antibiotics, and bacteria, contributing to the characterization of epidemic propagation in bacterial populations suffering different types of stress, and highlighting the role of the spatial structuration of the system.
31 January 2024 : François Rousset
What bacterial defenses teach us about innate immunity?
François Rousset, Postdoctoral scientist - Weizmann Institute of Science
Watch the replay here
Abstract
Pathogens have fueled the diversification of intracellular defense strategies that collectively define cell-autonomous innate immunity. In bacteria, innate immunity is manifested by a broad arsenal of defense systems that provide protection against phages. The complexity of the bacterial immune repertoire has only been realized recently and is now suggesting that innate immunity has commonalities across the tree of life: many components of eukaryotic innate immunity are found in bacteria where they protect against phages. Here, I summarize recent findings on the conservation of innate immune pathways between prokaryotes and eukaryotes. I show that bacterial defense mechanisms can in turn catalyze the discovery of novel molecular players of eukaryotic innate immunity. We recently described ATP nucleosidases, immune effectors which cleave ATP molecules into adenine and ribose-5'-triphosphate during phage infection, thereby depriving phages of energy. Using phylogenetic analyses, we found that the immune ATP nucleosidase domain is found in a variety of eukaryotic organisms ranging from fungi to corals and insects, where it is embedded in a diverse set of proteins with a typical immune architecture. Taken together, our findings suggest that ATP degradation represents a novel mechanism of innate immunity that is conserved across the tree of life, highlighting the potential of bacterial defenses to expand our knowledge of eukaryotic immunity.